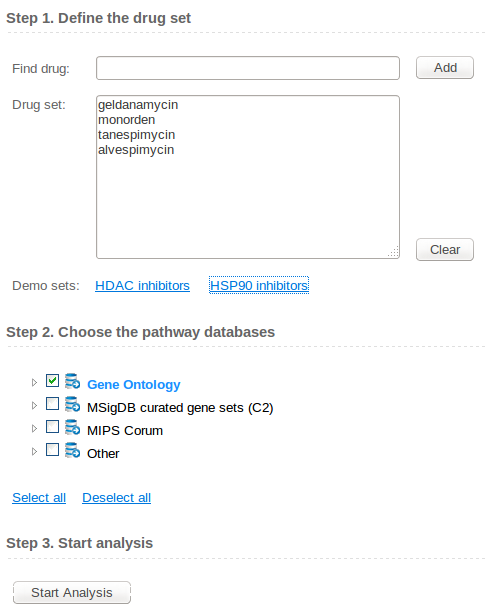
About
| ||
Napolitano, F., Sirci, F., Carrella, D. & di Bernardo, D. Drug-set enrichment analysis: a novel tool to investigate drug mode of action. Bioinformatics (2015). doi:10.1093/bioinformatics/btv536. | ||
| ||
|
A common problem when screening small molecules against a desired phenotypic change is that the screening hits can form an obscure, chemically heterogeneous set of molecules. | ||
| ||
|
As some examples of factors contributing to hide the relevant molecular mechanisms in action, consider the following:
| ||
| ||
|
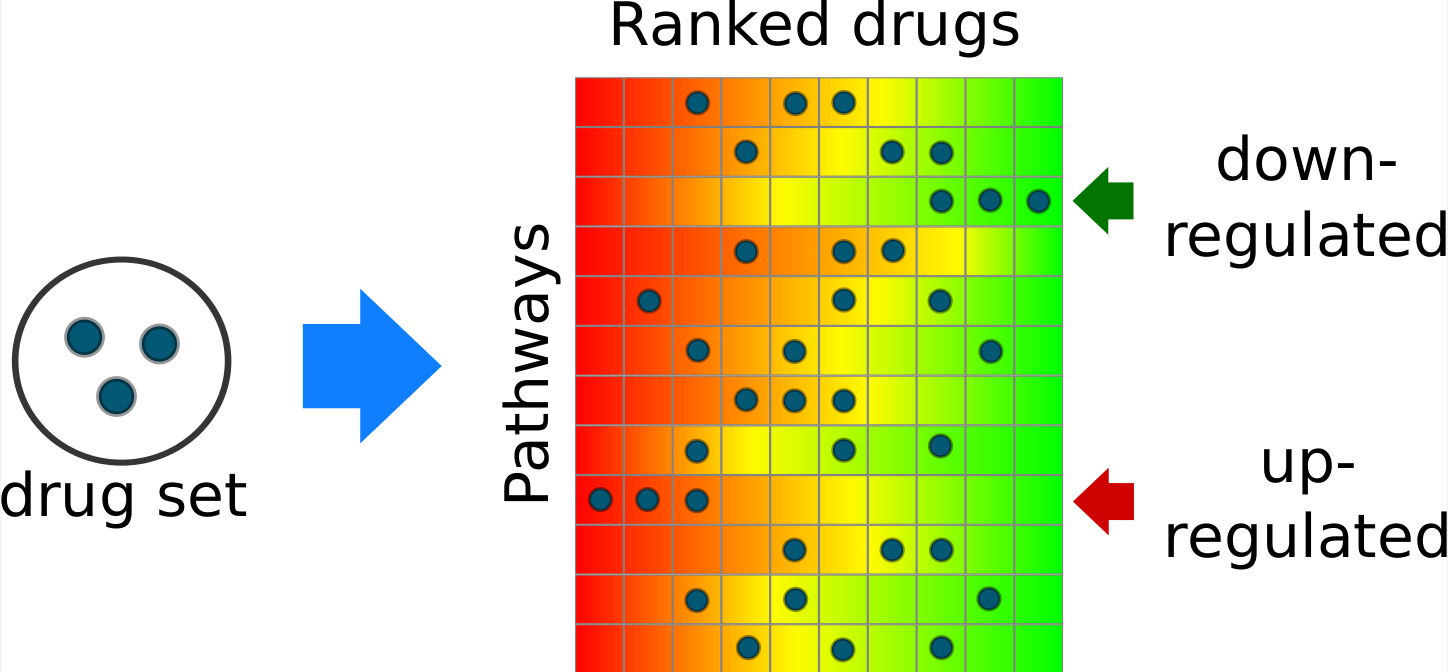
The Drug Set Enrichment Analysis (DSEA) works on the same principles as GSEA, but using an inverse preparation and interpretation of the data. A set of drugs of interest is tested against a database of pathways. Each pathway in the database is stored as a ranked list of drugs (Perturbagen Expression Profile), sorted from the one most up-regulating the pathway (or gene) to the one most down-regulating it. Current data is based on the Cmap 2.0. | ||
| ||
|
DSEA tend to highlight pathways that are most dysregulated by a set of drugs of interest compared with the other drugs in the database. The importance of a drug-pathway pair is not based on the absolute dysregulation level of the pathway induced by the drug, but rather on the ability of the drug to dysregulate the pathway more than other drugs. This could be the key behind the efficacy of the screening hits and the inefficacy of the other drugs. | ||
| ||
|
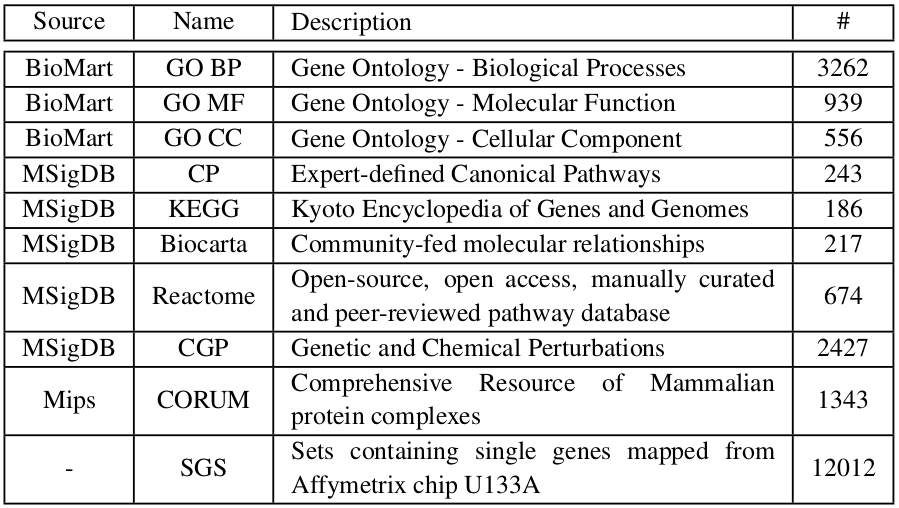
Pathways analyzed by the DSEA are defined as gene sets collected by various sources, as summarized in the following table: | ||
| ||
| ||
| ||
| ||
|
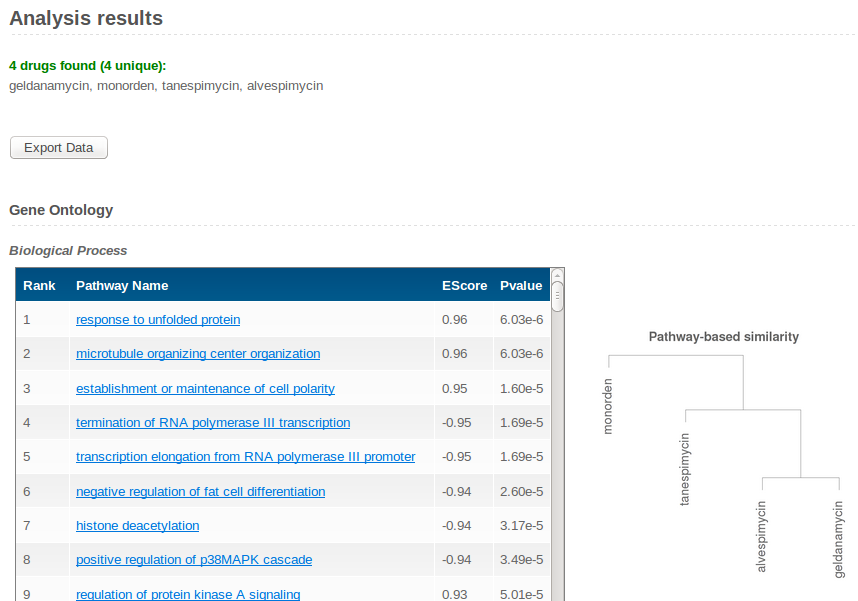
Results for each database are shown as separate tables, in which pathways are sorted according to relevance, together with the corresponding Enrichment Scores (ESs) and nominal p-values. Top 10% pathways are shown for each database. | ||
| ||
|
P-values assess how much the ranks of the chosen drugs are consistently small (up-regulation) or large (down-regulation) for each pathway. Positive ESs correspond to up-regulated pathways, negative ESs correspond to down-regulated pathways. Note that, due to cell adaptation effects, the direction of dysregulation could be opposite to that directly induced by the drug.
| ||
|
| ||
|
Use of DSEA tool is free to academic, government and non-profit users for non-commercial use. Use of DSEA application for commercial use by non-profit and for-profit entities requires a license agreement. Parties interested in commercial use may first obtain a demo version of a stand-alone version of DSEA application, subject to this License, by contacting Diego di Bernardo (dibernardo_at_tigem.it). Redistribution and use in source and binary forms, with or without modification, are permitted provided that the following conditions are met:
|